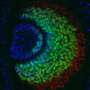
Gene cascade specifies two distinct neuron sets expressing Nplp1
A study of the embryonic nervous system of the fruit fly throws light on how two neuronal cell lineages that develop at different times and in different places in the ventral nerve cord of the embryo can ultimately result in very similar neuronal subtypes. The study, publishing in the Open Access journal PLOS Biology on 5th May, is a collaboration between research teams in Madrid (Spain) and Linköping (Sweden).
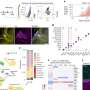
In the paper, Hugo Gabilondo, Johannes Stratmann and their colleagues report that a crucial terminal selector gene, col, is activated by different sets of spatio-temporal selector genes in the two different neuronal cell lineages. In dAp neurons, which are present throughout the thorax and abdominal segments, col is activated directly by the action of the early temporal genes Kruppel (Kr) and pdm, and the GATA transcription factor gene grain (grn). By contrast, in Tv1 neurons, which are specific to the thoracic segments, col is activated by the late temporal gene cas, together with several other genes that feed forward onto the terminal selector gene cascade downstream of col. The result is expression of the neuropeptide Nplp1 in both dAp and Tv1 neurons.
The developing nervous system generates many different neuronal cell types; understanding this process of cell fate specification remains a major challenge for biologists. Complex cascades of regulatory genes are known to be involved, starting with spatial and temporal selector genes and finishing with terminal selector genes, all of which act in various combinations to dictate the ultimate neuronal cell type. A particular neuronal cell type often arises in several parts of the nervous system and at different stages of development, however, suggesting that different spatio-temporal cues can converge on the same terminal selectors to generate a similar cell fate. This study reports evidence of this phenomenon in an example from the fruit fly, Drosophila melanogaster.
Previous work had shown that expression of Nplp1 in Tv1 and dAp neurons depends upon a single terminal selector gene cascade beginning with col. The progenitors of Tv1 cells were already well defined and several genes upstream of col were known to impinge on the terminal selector cascade in these cells. By contrast, the progenitor cells that give rise to dAp neurons were not known. By using sets of markers that identify progenitors of neuronal cells in the fly embryo, the Spanish and Swedish teams were able to identify the progenitor lineage that gives rise to dAp neurons, to show that that this lineage is entirely unrelated to that of the Tv1 neurons, and that it expresses distinct genes and has a different proliferation mode.
Then, by using gene expression analysis in mutant and misexpression backgrounds combined with cross-rescue and combinatorial misexpression studies, the authors determined that dAP neurons depend upon the early temporal genes Kr and pdm for their specification, whereas Tv1 neurons, which arise later, depend upon the late temporal gene cas. They also showed that grn is an early spatial cue acting upstream of col in the dAp specification cascade and that the Kr and pdm genes act further upstream to activate grn. The authors think it likely, however, that they are still missing additional upstream and feed-forward regulators to explain the highly localized, specific triggering of col in the dAp lineage.
Strikingly, the researchers found that grn is not involved in triggering the terminal selector cascade in Tv1 neurons. By contrast, another gene, ladybird early (lbe), identified in a previous screen for genes involved in Tv4 neuron specification, is required for Col and Nplp1 expression in Tv1 cells. lbe acts in parallel with the other spatiotemporal cues in this lineage in a feed-forward loop, first activating col and subsequently acting with col to activate the rest of the cascade.
Such feed-forward loops are common in bacteria and yeast gene regulatory networks and they have been identified during nervous system development in the nematode worm Caenorhabditis elegans. They act as regulatory timing devices that allow a single gene, such as col in this case, to have different regulatory outputs at successive times in development. The feed-forward loop identified here, incorporating five steps between Kr and Nplp1 in dAp cells, and ranging in developmental time from mid to late embryonic stages, is exceptionally long.
More information: Gabilondo H, Stratmann J, Rubio-Ferrera I, Millán-Crespo I, Contero-García P, Bahrampour S, et al. (2016) Neuronal Cell Fate Specification by the Convergence of Different Spatiotemporal Cues on a Common Terminal Selector Cascade. PLoS Biol 14 (5): e1002450. DOI: 10.1371/journal.pbio.1002450