Team finds key to tuberculosis resistance
The cascade of events leading to bacterial infection and the immune response is mostly understood. However, the molecular mechanisms underlying the immune response to the bacteria that causes tuberculosis have remained a mystery—until now. Researchers at the Johns Hopkins University School of Medicine have now uncovered how a bacterial molecule controls the body's response to TB infection and suggest that adjusting the level of this of this molecule may be a new way to treat the disease. The report appears this week as an advance online publication of Nature Medicine.
"We unraveled part of the cat-and-mouse game that plays out when TB bacteria infect human cells. The microbes release a small piece of DNA that resembles viral DNA, and this tricks the human cells to react as if they were responding to a virus instead of a bacterium; this may explain in part why the human immune response is often unable to combat TB," says William Bishai, M.D., Ph.D., co-director of the Johns Hopkins Center for Tuberculosis Research Laboratory and corresponding author on the paper. "The exciting part is that with the knowledge of this molecular trickery, we may be able to come up with better drugs and vaccines for TB—tools that are sorely needed."
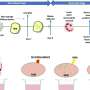
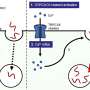
After tuberculosis bacteria infect a host, the bacteria release a molecule called c-di-AMP into the host's cells. Those cells have built-in early detection systems that recognize these foreign molecules and start an inflammatory response, which then leads to a complex reaction to combat the infection. The research team first measured c-di-AMP levels in the bacteria and found that its levels increase when the bacteria are actively multiplying.
To determine if c-di-AMP is indeed altering the host immune response, the researchers infected mouse immune cells with TB bacteria engineered to make different levels of c-di-AMP and compared how much of an immune response the cells mounted by measuring levels of INF-beta protein. They found that the more c-di-AMP released into the mouse cell, the higher the INF-beta levels.
But, according to Bishai, INF-beta levels may not reveal the whole picture of what transpires during infection. So they then looked at how well the bacteria themselves grow when releasing different amounts of c-di-AMP into the cells they've infected. The bacteria making the highest levels of c-di-AMP, it turns out, showed the slowest growth rates.
"Others had suggested that molecules of the same class as c-di-AMP can trigger autophagy, when a cell chews up and disposes of its insides," says Bishai. "So we set out to see if overproducing c-di-AMP was causing the infected host to eat the TB bacteria. Using cells marked with glowing proteins, the researchers saw under microscopes that cells infected with TB bacteria making high levels of c-di-AMP indeed underwent more autophagy than those with lower levels of c-di-AMP.
The team then examined whether differences in c-di-AMP could alter the severity of the disease in mice. Infection with normal bacteria causes death at about 150 days, whereas infection with bacteria engineered to overproduce c-di-AMP led to longer survival times—321 days.
"We still don't know if altering c-di-AMP levels can be linked to different outcomes in humans with TB, but this study does suggest that it would be well worth looking into," says Bishai.
More information: A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis, Nature Medicine, DOI: 10.1038/nm.3813