Neural protein mutations cause extreme brain malformations and atrophy
A gene responsible for a severe form of the brain formation disorder microlissencephaly has been identified by A*STAR scientists through a collaboration with researchers in seven countries.
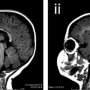
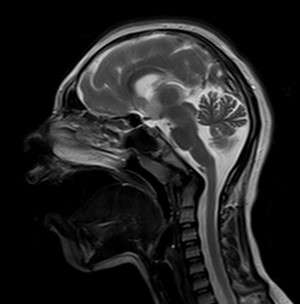
Microlissencephaly is a neuronal development disorder characterized by smaller brains having fewer neurons than normally developed brains (see image). Researchers, including Bruno Reversade from the A*STAR Institute of Medical Biology, identified three Middle Eastern families with children affected by the disorder who did not have mutations in any of the genes already known to cause microlissencephaly. Because some of the families had a history of marriages between closely related individuals, such as first cousins, the causative gene was believed to be passed on by both parents. By sequencing regions that were identical in the affected individuals of a particular family, the researchers were able to home in on mutations in the KATNB1 gene, as these mutations were absent in unaffected individuals in other populations.
The protein encoded by KATNB1 is part of a complex responsible for severing structural proteins in the cell called microtubules. A structure called the centrosome is known to play a central role in organizing microtubules in the cell, and many centrosomal proteins have previously been linked to patients with microlissencephaly.
To determine the function of KATNB1 during development, Reversade and his colleagues generated mice and fish lacking the Katnb1 gene—the mouse and fish analogue of KATNB1. The animal models showed defective proliferation of neuronal progenitor cells, loss of many cell types that rely on normal cell division, enhanced cell death and thinner brains. Also, cells derived from humans with microlissencephaly showed many abnormalities in cell division, such as centromeres that were abnormally placed, chromosomes that did not line up properly and cells with too many chromosomes.
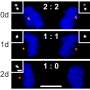
Centromeres are made up of paired structures called centrioles and play a key role in forming hair-like extensions of the cell called cilia. The researchers found that cells from mice lacking Katnb1 contained many more centrioles than normal cells and that these centrioles were often unpaired. Moreover, the cells lacking this gene contained many more cilia than normal, which the researchers argue could mean that signaling regulated by cilia is defective. These dysfunctional processes probably contribute to the abnormal neuronal development observed in organisms lacking KATNB1.
"Our discovery of the causative gene for this disease will benefit families as we are now able to provide premarital and prenatal diagnosis for parents," explains Reversade.
More information: "Katanin p80 regulates human cortical development by limiting centriole and cilia number." Neuron 84, 1240–1257 (2014). dx.doi.org/10.1016/j.neuron.2014.12.017