Fighting MRSA with new membrane-busting compounds
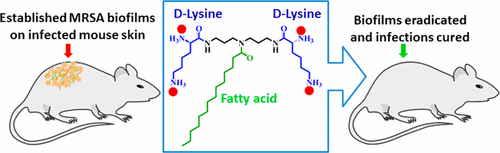
Public health officials are increasingly concerned over methicillin-resistant Staphylococcus aureus (MRSA). The bacteria have developed resistance to a number of treatments, even antibiotics of last resort in some cases. Now researchers report in ACS' journal Bioconjugate Chemistry that a new class of compounds can treat MRSA skin infections in mice with no signs of acute toxicity, and no signs that the bacteria would develop resistance to them after many applications.
According to the U.S. Centers for Disease Control and Prevention, every year at least 2 million people in America become infected by bacteria resistant to antibiotics, and 23,000 people die from such infections. Researchers have been working to combat this major public health threat for years. One of the latest fronts in this fight involves antimicrobial peptides and lipopeptides, which can destroy bacterial membranes. But translating these molecules into clinical products has been difficult. More recently, researchers have developed a new class of membrane-busting compounds called lysine-conjugated aliphatic norspermidine analogues (LANAs) that have been effective at killing certain bacteria and the Ebola virus in lab tests. Mohini Mohan Konai and Jayanta Haldar wanted to see if these compounds could also work against MRSA.
The researchers found that LANAs were effective against four MRSA strains in lab experiments. Testing on mice showed that the compounds could eliminate MRSA skin infections, which form notoriously difficult-to-treat biofilms. Even after 20 passages, the MRSA bacteria failed to develop resistance to the compounds. The results suggest that LANAs could be strong contenders for treating MRSA skin infections, the researchers say.
More information: Mohini M. Konai et al. Fatty Acid Comprising Lysine Conjugates: Anti-MRSA Agents That Display In Vivo Efficacy by Disrupting Biofilms with No Resistance Development, Bioconjugate Chemistry (2017). DOI: 10.1021/acs.bioconjchem.7b00055
Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) has developed resistance to antibiotics of last resort such as vancomycin, linezolid, and daptomycin. Additionally, their biofilm forming capability has set an alarming situation in the treatment of bacterial infections. Herein we report the potency of fatty acid comprising lysine conjugates as novel anti-MRSA agents, which were not only capable of killing growing planktonic MRSA at low concentration (MIC = 3.1–6.3 μg/mL), but also displayed potent activity against nondividing stationary phase cells. Furthermore, the conjugates eradicated established biofilms of MRSA. The bactericidal activity of d-lysine conjugated tetradecanoyl analogue (D-LANA-14) is attributed to its membrane disruption against these metabolically distinct cells. In a mouse model of superficial skin infection, D-LANA-14 displayed potent in vivo anti-MRSA activity (2.7 and 3.9 Log reduction at 20 mg/kg and 40 mg/kg, respectively) without showing any skin toxicity even at 200 mg/kg of the compound exposure. Additionally, MRSA could not develop resistance against D-LANA-14 even after 18 subsequent passages, whereas the topical anti-MRSA antibiotic fusidic acid succumbed to rapid resistance development. Collectively, the results suggested that this new class of membrane targeting conjugates bear immense potential to treat MRSA infections over conventional antibiotic therapy.
Journal information: Bioconjugate Chemistry
Provided by American Chemical Society