Atlas of immune cells shows unexpected complexity, but could ultimately offer valuable clinical insights
T cells help coordinate the immune response against both infectious threats and tumors. However, this is not merely a single class of cells, but a diverse population with specialized functions and properties. A*STAR researchers have now conducted a detailed T cell census to reveal the extent of this diversity, which may inform better diagnosis and treatment of human disease.
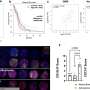
T cells are typically classified by the proteins they display on their surface that allow cells to respond to specific signals in their environment. A Singapore-based team led by Evan Newell and Michael Wong of the A*STAR Singapore Immunology Network had previously profiled T cells in the bloodstream using a technology called mass cytometry. This made it possible to classify cells based on many different protein markers in parallel.
"However, we realized this technique was only giving us a view of a small fraction of the various types of T cells in humans," says Newell. T cells can be found in many different tissues of the body, and a subset of the proteins on their surface act as 'trafficking receptors' that help recruit specific cell types to their appropriate destination.
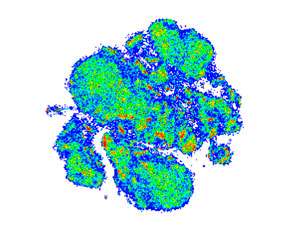
Newell, Wong and colleagues therefore embarked on a more extensive analysis, in which they tracked 41 different cell-surface markers in samples from eight different human tissues. Their hope was to identify distinctive sets of markers that represent 'address labels' for the various T cell subtypes.
Their pursuit paid off and the researchers were able to construct multiple different profiles based on distinct combinations of surface proteins. However, the biology proved more tangled than expected.
"The most interesting finding was also the most frustrating," says Newell. "T cell phenotypic and functional profiles are incredibly complex and difficult to neatly classify into easy to understand subsets." For example, several of the proteins appear to be associated with trafficking to many different destinations, and predictions of T cell function based on mouse data did not hold up in humans.
Nevertheless, this rich dataset is a powerful resource for clinical researchers. Many medical interventions rely specifically on a T cell-mediated response, including vaccines and cancer-targeting immunotherapies. Newell notes that this diversity could profoundly affect the success or failure of such approaches, and his team is now investigating the clinical importance of these T cell subsets. "We are now focusing on disease scenarios such as cancer and infectious disease to understand what happens to these T cells in those conditions," he says.
More information: Michael Thomas Wong et al. A High-Dimensional Atlas of Human T Cell Diversity Reveals Tissue-Specific Trafficking and Cytokine Signatures, Immunity (2016). DOI: 10.1016/j.immuni.2016.07.007