Lineage tracing in the gut
Scientists are finding that they can understand organ function (and dysfunction in the case of disease) better if they know where the cells that make up these organs came from.
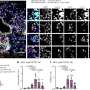
New single-cell technologies allow all cell states within a differentiating tissue to be identified. However, the relationships between cell states that lead to formation of tissues have been difficult to define from the large volumes of data generated.
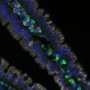
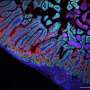
Now Ken Lau, Ph.D., and colleagues have overcome that challenge with an algorithm they developed called p-Creode. When applied to single-cell RNA-seq and imaging data of a poorly understood population of chemosensory cells in the gut called "tuft cells," p-Creode uncovered alternative routes of development in the small intestine and colon.
Because tuft cells play a role in immune responses against helminth (parasitic worm) infections, understanding and potentially modulating tuft cell development may be important in controlling inflammatory diseases in the gut, the researchers reported in the journal Cell Systems.
More information: Charles A. Herring et al. Unsupervised Trajectory Analysis of Single-Cell RNA-Seq and Imaging Data Reveals Alternative Tuft Cell Origins in the Gut, Cell Systems (2017). DOI: 10.1016/j.cels.2017.10.012