New research uncovers 'stability protein' for cancer treatment
Researchers from the Novo Nordisk Foundation Center for Protein Research at the Faculty of Health and Medical Sciences have characterised a new protein that is important to the genetic stability of cells. It may be significant for the development of new drugs against genetically determined diseases like cancer, sterility and premature ageing.
All cell nuclei contain genetic material, DNA, which controls the activity of the cells. If the genetic material is damaged, cancer cells may develop. Therefore, many proteins and enzymes are responsible for stabilising and protecting our DNA against permanent damage and mutations.
Researchers from the Novo Nordisk Foundation Center for Protein Research at the Faculty of Health and Medical Sciences, University of Copenhagen, have discovered and characterised a new protein called ZUFSP. There is much indication that the protein plays a key role in ensuring that our genetic material remains stable.
"The protein ZUFSP had not been characterised before, but appeared to contain certain sequences often found in proteins involved in what is referred to as DNA damage response. In addition, there is much indication that ZUFSP plays a main role in helping the cells maintain genetic stability. If we remove ZUFSP, the cells become genetically unstable," says the head of research, Professor Niels Mailand.
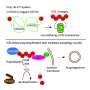
Genetic stability plays a key role in various aspects of human health. Genetic instability leads to cancer, neurodegeneration, immunodeficiency, sterility and early aging, among other conditions. The body's DNA damage response ensures that DNA is repaired correctly. It acts as an alarm system that responds every time damage or change occurs to DNA. Among other things, the damage response sends specific molecules to the damage site. One of these molecules is called ubiquitin, which is connected in chains and can act as a directory signal for the proteins responsible for repairing the damage. Ubiquitin can also stop cell growth until the damage or error has been repaired.
If it proves impossible to repair extensive DNA damage, the cell is usually programmed to destroy itself. However, due to genetic changes caused by failed DNA repair, cancer cells do not always do so. Instead, the cancer cell splits up, creating many new cells, which can potentially develop into a cancer tumour.
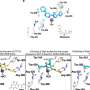
The newly discovered protein belongs in a class of enzymes called deubiquitylating enzymes or DUBs. Their function is to remove the ubiquitin chains, once they have done their job.
"Besides ZUFSP appearing to play a role in genetic stability, we also unexpectedly discovered that ZUFSP is a DUB capable of removing a specific type of ubiquitin chain. This type of ubiquitin chain is found in the area around DNA damage and attracts important repair proteins to the damage," says Mailand.
There are six known DUB classes. The newly discovered ZUFSP does not fit into any of these classes and therefore defines its own seventh class. The researchers still do not know exactly why it is important for ZUFSP to remove the ubiquitin chains from the area around DNA damage. They believe it is possible that balance plays a role with regard to repairing DNA damage. In the case of an imbalance between the proteins that attach the chains and the ones that remove them again, the proteins do not perform the repair as well as when they are in balance.
When the researchers first discovered the uncharacterised ZUFSP, they observed that to a large extent, it behaved like other proteins involved in DNA repair. A lot of these proteins accumulate physically around DNA damage. This is also the case for ZUFSP.
"When we wish to examine whether a protein like ZUFSP plays a role in DNA damage response, we can, for example, choose to attach a small marker to the protein called green fluorescent protein. It glows green under the microscope. If we then induce DNA damage to a cell, we can see that ZUFSP, now glowing green, physically moves towards the DNA damage," says Mailand.
ZUFSP's movement towards DNA damage suggests that the protein, in addition to removing ubiquitin chains, also plays a role in DNA damage response. So far, the researchers do not know exactly what that role is.
Even though more research is required, the discovery of ZUFSP is significant.
"This is a very basic discovery. Right now, we know what ZUFSP is and what it can do. Now we must try to understand what is happening at the molecular level and its possible biological relevance to pathogenesis," says Mailand.
With regard to disease, ZUFSP and other DUBs may eventually make a huge difference in the future. "There is a lot of focus on drug discovery on these DUBs. It is a new pioneer area with promising potential when it comes to developing cancer treatments and other drugs," he says.
The study, "ZUFSP Deubiquitylates K63-Linked Polyubiquitin Chains to Promote Genome Stability," has been published in Molecular Cell.
More information: Peter Haahr et al. ZUFSP Deubiquitylates K63-Linked Polyubiquitin Chains to Promote Genome Stability, Molecular Cell (2018). DOI: 10.1016/j.molcel.2018.02.024