Allen Institute for Brain Science database release nearly doubles mouse brain cell data
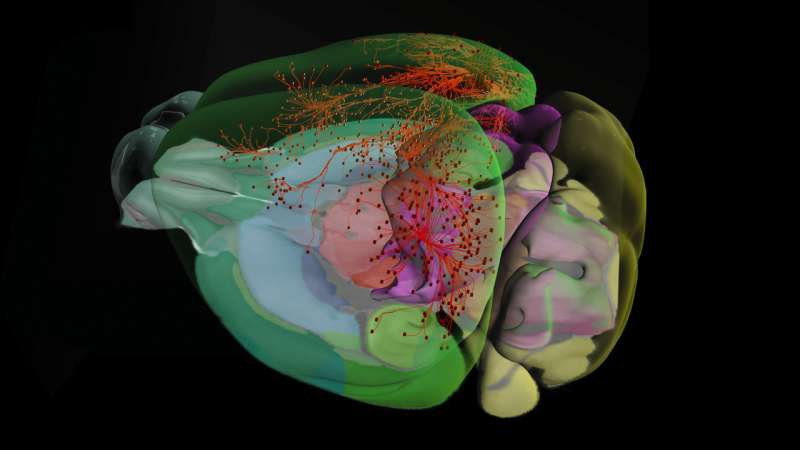
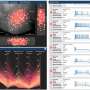
The Allen Institute for Brain Science today announced the release of new data, tools to analyze those data and a new web-based 3-D viewer to explore anatomy and connections in the mouse brain, the Allen Brain Explorer. Today's data release almost doubles the number of mouse cells in the Allen Cell Types Database—raising the number included from just over 1,000 to slightly more than 1,900 cells—as well as adding new neuronal connectivity information to the Allen Mouse Brain Connectivity Atlas and adding more single-cell transcriptomics data from mouse, human and macaque brain cells.
The growth of datasets contained in the Allen Brain Atlas allows researchers at the Allen Institute for Brain Science and in the broader neuroscience community to classify brain cells into different categories that help researchers understand more about the mammalian brain and its diversity, part of an ongoing effort to create a "periodic table" of brain cell types. As more data accumulate, those classifications provide a richer description of cellular complexity, said David Feng, Ph.D., associate director of technology at the Allen Institute for Brain Science.
"We use different aspects of the cells—their gene activity, their shape, where they send the information to, their electrical behavior—to come up with classifications that have biological meaning," Feng said. "Understanding the properties of cells in the brain is a critical step toward understanding how the brain works."
Because the brain is so complex, Allen Institute for Brain Science researchers need to collect large amounts of data from tens of thousands of brain cells to reliably sort the different cells into categories and begin to understand what those categories mean.
In addition to the new mouse brain cell data, today's release includes data at the single cell level from human and macaque brains, through collaborative projects with teams at UW Medicine and the Swedish Neuroscience Institute. These precious data sets provide researchers with insight into differences between the human and animal brains, which are used to model human diseases.
"Diseases that affect the brain may start from a specific part of the brain or even a specific cell type," said Bosiljka Tasic, Ph.D., associate director of molecular genetics at the Allen Institute for Brain Science. "To know where a disease starts—and ultimately how to treat it—we have to understand the individual building blocks of the brain."
Today also marks the beta release of two new data analysis tools. One, the Allen Brain Explorer, is a web-based application that allows users to visualize mouse brain anatomy and connectivity in 3-D. Previously, this tool had been available as a desktop application only; now, users can reach the explorer through their web browsers. The second tool, called RNA-Seq Data Navigator, allows researchers to visualize categories of brain cells based on their gene similarity patterns. These open data analysis applications join a rich toolkit for clinicians and neuroscientists around the world to use at no cost.