Researchers unlock the secrets of liver regeneration
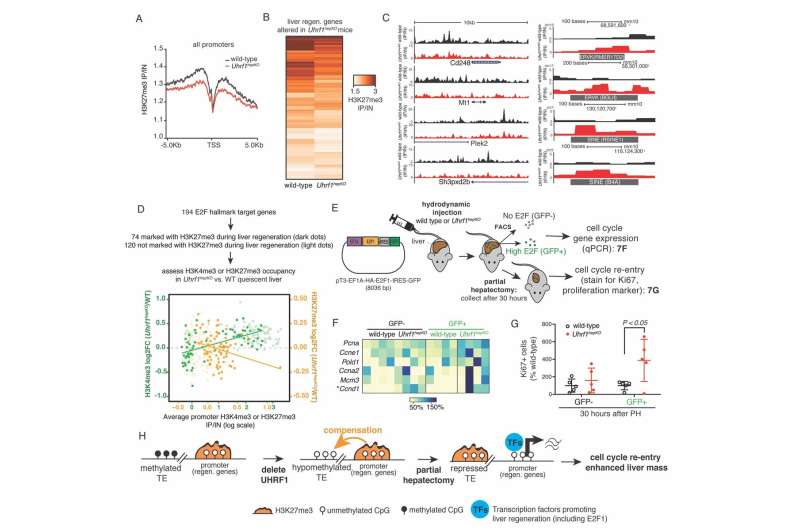
In a recent study published in the journal Developmental Cell, NYU Abu Dhabi researchers have reported a new way in which the liver is primed to regenerate itself. They found that by stripping parts of the epigenome, which play a primary role in repressing "jumping genes" (i.e. transposable elements), other epigenetic marks were redistributed.
This newly discovered form of epigenetic compensation protects the genome against transposable elements activation, but takes these compensating epigenetic marks away from their normal job in regulating gene expression. The result is that when these marks are taken away from their normal role, the genes they usually repress are activated early and are sustained during the regenerative response to the surgical removal of part of the liver.
This type of surgery is relevant to humans, as it is used in resection of liver tumors and the regenerative response is essential for the liver to respond to damage. The findings are a significant advance in the understanding of the liver regeneration process, which is unique among the organs of humans, mice, and other mammals.
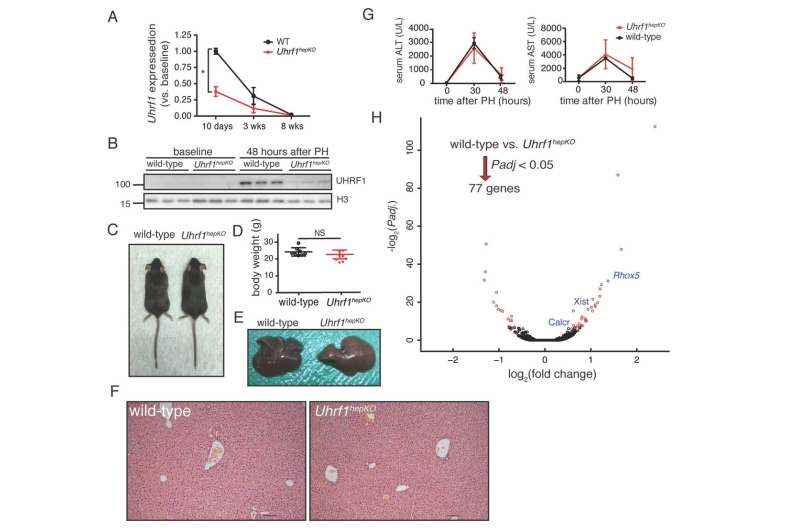
The researchers from NYU Abu Dhabi's Sadler Lab, led by Associate Professor of Biology Kirsten Sadler Edepli, removed a key epigenetic regulator, UHRF1 in the mouse liver. They found when they removed part of the liver, the remaining lobes responded more readily by activating pro-regenerative genes activated earlier, and this regeneration program stayed active longer, resulting in enhanced liver regeneration.
The epigenome refers to the code that packages the genome so that some parts can be activated (i.e. genes) and some parts remain in dormant domains—these dormant parts largely contain remnants of old viruses or transposable elements, which were made famous by the 1983 Nobel Prize discovery by Barbara McClintok.
Surprisingly, instead of causing massive activation of transposable elements or an immune response to mitigate the unleashing of transposable elements, as found in previous experiments, they discovered that there is an extra layer of protection by another repressive epigenetic mark (H3K27me3). This mark was redistributed from gene promoters to suppress transposable elements when DNA methylation was missing, thereby compensating for the loss of DNA methylation. When this mark is redistributed, it is removed from its role in repressing genes that promote liver regeneration. Thus, livers lacking UHRF1 are able to regenerate faster.
"When H3K27me3 compensates for the loss of DNA methylation, this results in a favorable epigenetic environment for liver regeneration," said Sadler Edepli. "It will be exciting to explore whether drugs that can modify the epigenome have the potential to induce epigenetic compensation and increase the liver's ability to regenerate in cases of liver disease or failure." Shuang Wang, a post-doctoral fellow in the Sadler Edepli laboratory who worked in her group at Icahn School of Medicine at Mount Sinai, led the study in collaboration with members of the lab at NYUAD as well as Emily Bernstein and Amaia Lujambio in NY.
More information: Developmental Cell (2019). DOI: 10.1016/j.devcel.2019.05.034 , www.cell.com/developmental-cel … 1534-5807(19)30447-2