Novel method identifies the right individual exosomes
There is a growing demand for diagnostic markers for early disease detection and prognosis. Exosomes are potential biomarkers for cancer progression and neurodegenerative disease, but it can be difficult to identify what tissue a specific exosome comes from. Researchers at Uppsala University and spin-off company Vesicode AB have solved this problem by developing a method that maps surface protein complements on large numbers of individual exosomes.
Exosomes are released from all cells in the body. They convey protein and nucleic acid cargos between the cells as a form of intercellular communication, and they represent potential circulating biomarkers for tumor progression and metastasis, as well as for early detection of neurodegenerative disease.
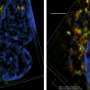
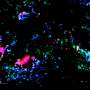
In order to use exosomes as biomarkers of diseases in different tissues it is vital to distinguish them according to their surface protein complements. Researchers at Uppsala University and Vesicode AB, along with collaborators, have developed a method that can map surface protein complements on large numbers of individual exosomes.
The novel proximity-dependent barcoding assay (PBA) reveals the surface protein composition of individual exosomes using antibody-DNA conjugates and next-generation sequencing. The method identifies proteins on individual exosomes using micrometer-sized, uniquely tagged single-stranded DNA clusters generated by rolling circle amplification.
"This technology will not only benefit researchers studying exosomes, but also enable high-throughput biomarker discovery. We will further develop and validate the PBA technology and provide service to researchers starting later this year. We believe single exosome analysis will allow this exciting class of biomarkers to reach its full potential," says Di Wu, researcher and inventor of the PBA technology and founder of Vesicode AB, commercializing the technique.
"This new technology will allow large-scale screens for biomarkers in disease, complementing a panel of methods for sensitive and specific detection of exosomes that we have previously established," says Masood Kamali-Moghaddam one of the group leaders at the Molecular Tools unit at Uppsala University.
The study is published in Nature Communications.
More information: Wu et al. (2019) Profiling surface proteins on individual exosomes using a proximity barcoding assay, Nature Communications, DOI: 10.1038/s41467-019-11486-1