Nervous system discovery overturns previous theory
It appears that when the nervous system is developing, only the most viable neurons survive, while immature neurons are weeded out and die. This is shown in a groundbreaking discovery by researchers at Karolinska Institutet in Sweden. The results indicate that the longstanding neurotrophic theory, which states that chance determines which cells will form the nervous system, needs to be revised.
During the early stages of the development of the nervous system, an excess of neurons is generated. At a certain time, a large portion of these cells then die, which is a necessary step for the proper formation of the nervous system. The process takes about 24 hours, and in certain parts of the nervous system, roughly half of all neurons disappear.
Researchers have previously believed this to be a random process in which all cells have an equal chance of survival. However, researchers from Karolinska Institute have now publishing a study in Nature Communications showing that cell death instead appears to be controlled by a mechanism that weeds out the less fit cells.
"The cells that survive are more mature and inclined to form synapses with other nerve cells," says Saida Hadjab, who coordinated the study together with Francois Lallemend. They are both researchers at the Department of Neuroscience at Karolinska Institutet.
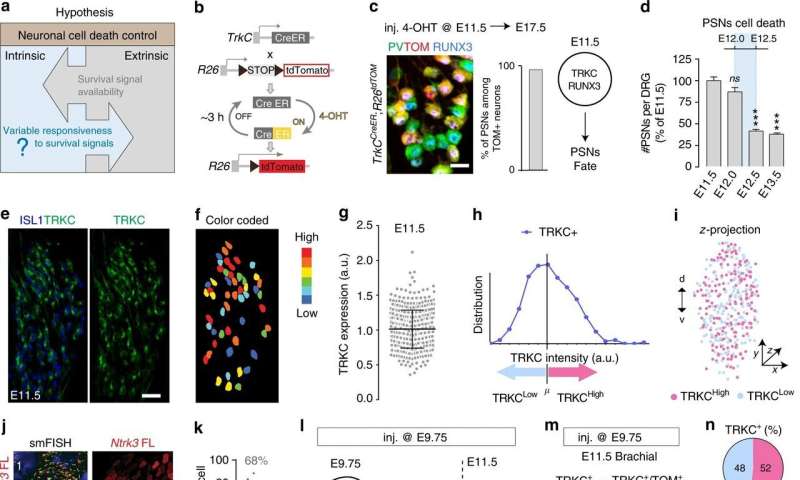
Several years ago, Hadjab and Lallemend noted that the early neurons are different. On their surfaces are receivers of growth factors that stimulate their survival. Hadjab and Lallemend discovered that certain neurons have more of these receivers than others. They started to suspect that cell death is somehow controlled such that only certain selected cells survive.
The researchers have now conducted a detailed study of individual neurons in the early nervous system in mice, and among other things, have discovered which genes are active. Their mapping revealed two distinct molecular patterns that determine the fate of these cells. The cells that are the most capable of growing and forming connections to other neurons survive, while the more immature cells die.
The study was performed in the peripheral sensory nervous system. Whether cell death is controlled in the same way in other parts of the nervous system remains to be discovered.
"This discovery can help us understand the brain and the development of the nervous system on a different level. Earlier studies have mainly studied the environment surrounding the cell, and neuronal population as homogenous cell type. Researchers have not examined the actual neurons individually, their fitness and how different they are," says Francois Lallemend.
This discovery could potentially be significant to the treatment of different neurological diseases. For example, in the case of Parkinson's disease, doctors have tried to transplant healthy stem cells in patients, but the majority of cells die shortly after the treatment. It is possible that the treatment could become more successful if the less fit cells were weeded out before the transplant so that the patient was only given viable neurons.
More information: "A cell fitness selection model for neuronal survival during development", Yiqiao Wang, et al Nature Communications, September 12, 2019, DOI: 10.1038/s41467-019-12119-3. www.nature.com/articles/s41467-019-12119-3