Team plucks needle from genomic haystack, finding essential transcription factor binding sites
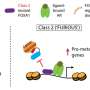
Using CRISPR/Cas9 knockout screens a multi-institutional research team systematically interrogated the essentiality of more than 10,000 forkhead box protein A1 (FOXA1) and CTCF binding sites in breast and prostate cancer cells, plucking useful needles from a massive genomic haystack that contains millions of transcription factor binding sites. They found that essential FOXA1 binding sites act as enhancers to orchestrate the expression of nearby essential genes, the team reports Nov. 11, 2019, in PNAS.
"Ninety-nine percent of the human genome is non-coding DNA, which previously had been thought of as junk," says Wei Li, Ph.D., a principal investigator in the Center for Genetic Medicine Research at Children's National Hospital and co-lead study author. "We now know that the non-coding regions of the genome can play important roles in a lot of biological functions, including cancer cell growth. The problem is there was no good way to figure out which among the millions of candidates are important in the biology of cancer."
While previous techniques interrogated a few hundred non-coding genomic regions, Li says their team was able to test more than 10,000 sites in a single experiment.
Overall, the team found 37 FOXA1 binding sites in T47D cells are essential, including 29 strong FOXA1 binding sites and eight binding sites near essential genes. That includes estrogen receptor 1, "the master transcription factor for ER+ breast cancer cells," and TRPS1, another transcription factor associated with ER+ breast cancer progression, the research team writes.
Li says the most exciting part of the work is the machine learning model they developed to predict which potential transcription binding sites are most important, yielding clinically relevant information that in the future may help patients.
"We have only finished the first step. We need to improve our machine-learning model. We need to conduct many more experiments. We need to test on cell lines using experimental models. And, we eventually hope to launch clinical trials to validate our findings in humans," he says. "It will be years from now, but we hope our machine learning model can one day be used to tell a patient which of the variants located in their genome may affect their risk of getting cancer."
More information: Teng Fei el al., "Deciphering essential cistromes using genome-wide CRISPR screens," PNAS (2019). www.pnas.org/cgi/doi/10.1073/pnas.1908155116