Researchers identify mechanism that triggers a rare type of muscular dystrophy

A study led by the IBB-UAB has identified the molecular mechanism through which a protein carrying genetic mutations associated with limb-girdle muscular dystrophy type 1G accelerates its tendency to form amyloid fibrils and triggers the appearance of the disease. The research, published in Cell Reports, will pave the way for the study of possible treatments.
Limb-girdle muscular dystrophy (LGMD) is the term given to a group of rare hereditary diseases characterised by the wasting and weakening of the hip and shoulder muscles. LGMD type 1G (LGMD1G) is associated with two possible genetic mutations in a protein called hnRNPDL. Little is known about this protein, except that it exists in cells in three functional forms (isoforms) and that it may contain the genetic mutations linked to the disease.
A research team led by the Institute of Biotechnology and Biomedicine at the Universitat Autònoma de Barcelona (IBB-UAB) now explains the behaviour of this protein, its role in the cells and the phenotype caused by the genetic mutations associated with LGMD1G, in an article published in Cell Reports.
The research establishes that one of the protein's isoforms demonstrates a greater tendency to form amyloid fibrils—toxic protein aggregates—and this tendency to aggregate occurred significantly faster when the protein contained the genetic mutations related to the disease, which prevented it from performing correctly.
"For the first time, we can provide solid proof of the effects genetic mutations have on the process of the hnRNPDL protein aggregation," says Salvador Ventura, IBB-UAB researcher and coordinator of the study. "Based on data obtained with the Drosophila fruit fly, we were able to suggest a possible mechanism for the disease: that it is the loss of protein function, once the aggregates are formed, that triggers the dystrophy. A hypothesis corroborated by the first data we are beginning to obtain with humans, and that opens the door to search for possible treatments."
Differential Behaviour
To conduct the study, researchers first analyzed the presence and behavior of the three isoforms in which the protein is found within the cells: with three, two or one protein domains, or independent regions. Then they studied the effects of the genetic mutations in the most common variant.
The isoform with two domains is most common in cells and, surprisingly for researchers, is also the one with the greatest tendency to form aggregates.
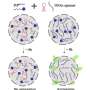
The researchers also saw that the isoform with three domains has a greater tendency to undergo a process known as phase separation, discovered a few years ago and of great biological importance, which could act as a prevention against the aggregations.
"What we have seen is that the more tendency towards phase separation, the less aggregates are formed. Until now, it was thought that phase separation was a process occurring after amyloid-type aggregation, and we have now seen that it is not always so," explains Salvador Ventura.
The study was conducted both in vitro and in human cells. It was also conducted on a transgenic model of the Drosophila fruit fly, in which the flies expressed their natural variant or each of the forms associated with the disease.
More information: Cristina Batlle et al. hnRNPDL Phase Separation Is Regulated by Alternative Splicing and Disease-Causing Mutations Accelerate Its Aggregation, Cell Reports (2020). DOI: 10.1016/j.celrep.2019.12.080