January 26, 2015 feature
Live long and prosper? Decoupling lifespan and healthspan in aging research
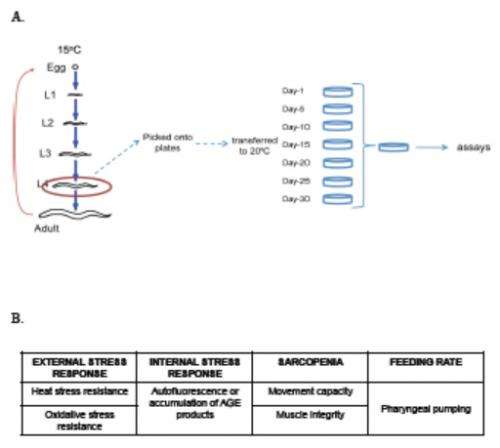
(Medical Xpress)—A long-standing assumption in aging research, and therefore in resulting antiaging interventions, is that modifying genetic and environmental factors to extend lifespan also increases healthspan – the length of time an organism is in optimal health. As it turns out, however, that assumption has escaped scientific scrutiny due largely to its apparent validity. Recently, scientists at University of Massachusetts Medical School (UMMS) examined this assumption, and thereby the life-/healthspan relationship, by investigating multiple lifespan-modulating signaling pathways – mechanisms in which ligands (ions or molecules) bind to receptors, resulting in altered cellular responses – in the wild type and four long-lived mutants (insulin/insulin-like growth factor-1, dietary restriction, protein translation, and mitochondrial signaling) of the roundworm Caenorhabditis elegans, a species studied for over two decades in aging research. They found that all long-lived mutants spent a higher percentage of their extended lifespan in a frail, rather than healthy, state. The scientists therefore conclude that aging research must include additional factors beyond lifespan alone.
Dr. Heidi A. Tissenbaum discussed the paper that she, Dr. Ankita Bansal and their colleagues published in Proceedings of the National Academy of Sciences. "C. elegans aging research has identified multiple pathways that have been shown to modulate lifespan," Tissenbaum tells Medical Xpress, This means that that there are individual genes that one can either increase or decrease in dosage, thereby leading to changes in the animal's lifespan – which, similar to humans, is measured in the number of days the animal lives. The researchers tested four mutants with mutations in the following genes, where each mutant displays an increased lifespan relative to wild type roundworms:
1. daf-2, the C. elegans insulin/IGF-1 receptor, where mutants show a doubling of lifespan
2. eat-2, a nicotinic acetylcholine receptor subunit routinely used as a genetic model of dietary restriction, with mutants showing a 33% increase in mean lifespan
3. clk-1, an enzyme necessary for biosynthesis of ubiquinone (a component of the electron transport chain required in aerobic cellular respiration, generating energy in the form of ATP), was the model for mitochondrial activity modulation, with mutants having a 25% increase in mean lifespan
4. ife-2, the gene encoding for the C. elegans eukaryotic initiation factor 4E, a principal regulator of protein synthesis, where mutants had a 40% increase in mean lifespan
The main challenge the scientists encountered was how to define healthspan – for example, as healthy aging – and moreover, how to define healthspan in the laboratory using C. elegans. "We read the literature, had many lab discussions and came up with the experimental framework that we used in our manuscript," Tissenbaum recounts. "We reasoned that we wanted to model human health, so from the literature and discussions, we concluded that we had to model both frailty and the ability to maintain homeostasis" – that is, resistance to external stress. "Next, the main challenge in the multiple cellular and organismal assays we conducted was the large number of animals we needed for the assays and the time required to carry out all of the experiments."
Time, indeed: The experiments described in the paper were performed entirely by Dr. Bansal – and it took several years. "We also challenged ourselves, other lab members, and other UMMS faculty to determine how to best illustrate the data," Tissenbaum adds. "Even the figures in the paper are the result of many hours discussing how best to illustrate both the change in a health parameter as well as the lifespan analysis in the same figure." (Note that the paper's Acknowledgements section provides an extensive list of research technicians, graduate students, post-doctoral fellows, and faculty who contributed to these discussions.)
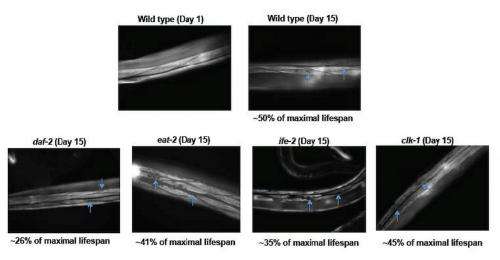
Interestingly, Tissenbaum tells Medical Xpress, showing that lifespan and healthspan can be separated was not expected. "This was quite surprising for us. We'd assumed that lifespan and healthspan would be correlated based not only on our data that many of the long-lived mutants showed the ability to resist external stress, but also on the fact that many papers use the word healthspan with limited assays while vaguely defining what healthspan meant."
The same held true for all of the long-lived mutants extending the period of frailty. "This was also surprising," she continues. "Prior to our work, we thought that there could be some costs to lifespan extension given prior studies as well as evolutionary theories. However, we didn't think we'd see the effects that we observed when we analyzed the raw data. In fact, personally, it was a bit disappointing: a main aspect of my thesis work – in Dr. Gary Ruvkun's laboratory at Harvard University – was identifying and characterizing the daf-2 gene, which I really wanted to be the gene where mutants showed both lifespan and healthspan extension."
Tissenbaum notes that the key insights from the study arose from the scientists conducting a broad study where they tested multiple parameters of health as the animals aged, comparing the four long-lived mutant types to wild type in every assay as the animals aged. "Another novel aspect of our study was that we analyzed the animals up to 80% of their average lifespan – so if one strain lives an average of 20 days, we tested up to 16 days, while for daf-2, where the average is 45 days, we tested up to 36 days. Next, when we analyzed the data, we did it in two ways: first by simply looking at the number of healthy days versus frail, and then analyzing the data based on the average lifespan." For example, she illustrates, an analysis at 50% of the average life span would differ in the animals mentioned above – that is, 10 days versus 22.5 days.
In addition to the mouse studies mentioned in the paper, Tissenbaum notes that additional mammalian studies may be conducted. "We revealed new insights largely because we took the broad approach of testing multiple assays as the animals aged, as well as testing the animals late into their lifespan. Similar experiments where mammals are assayed late into life, with healthspan and gerospan being measured chronologically and physiologically, might be performed in the near future. Looking further ahead," she continues, "more extensive studies dissecting healthspan and lifespan in mammals would be exciting. The biggest challenge would be the cost of maintaining a large number of older animals. Of course, intensive studies in people would be a goal for future work but this would require first defining health, healthspan and parameters to test in people."
Moving forward, the scientists are interested in identifying genes and interventions that extend healthspan and compress gerospan. "We've set up a perfect system to test this, and are developing new methods to automate some of the assays." The researchers will be seeking additional funding to continue and expand their work – and also welcome collaborators.
While synthetic biology provides techniques for altering/synthesizing genes in order to modify expression into phenotype, Tissenbaum sees these areas as far removed from what the researchers do in their lab. "In thinking about our work, aging is a whole body process. Our statistical modeling shows no single equation could account for the decline in more than one health parameter, suggesting that the rates of decline are different and indicates that – in addition to hormonal crosstalk between tissues – there's probably tremendous tissue specificity in the aging process."
"Ultimately, all medical research is aimed at prolonging life – but we must strive to prolong healthy life. Therefore, our challenge remains defining health and the ability to test and quantify healthy living. For human studies," Tissenbaum concludes. "I believe studies involving extensive databases of family history through generations – such as the Framingham Heart Study and the Kuakini Hawaiian Lifespan Study – will be key."
More information: Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants, Proceedings of the National Academy of Sciences (2015) 112(3):E277-E286, doi:10.1073/pnas.1412192112
© 2015 Medical Xpress


















